Damien O'Halloran

Damien O'Halloran
Associate Professor of Biology
Cellular & Molecular Biology
Contact:
My lab is interested in how nematodes sense their environment. Parasitic nematodes constitute one of the major threats to human health by causing diseases of major socio economic importance. Recent estimates indicate that more than 1 billion people are infected with parasitic nematodes, and more than a dozen species routinely parasitize humans. To address these questions we use the model nematode, Caenorhabditis elegans, to understand how animals detect and encode sensory information. By using the more compact C. elegans nervous system to address these questions we can leverage powerful genetic tools and anatomical datasets to dissect these problems. We also utilize parasitic nematodes (unlike C. elegans, which is free-living) that deploy sensory systems to find their specific host(s) and recognize when they are inside the correct host. For these studies we use the insect parasitic nematode, Heterorhabditis bacteriophora, and the intestinal blood-feeding species of hookworm. We have recently developed a database of nematode chemoreceptors, which are critical receptors for host specific cues.
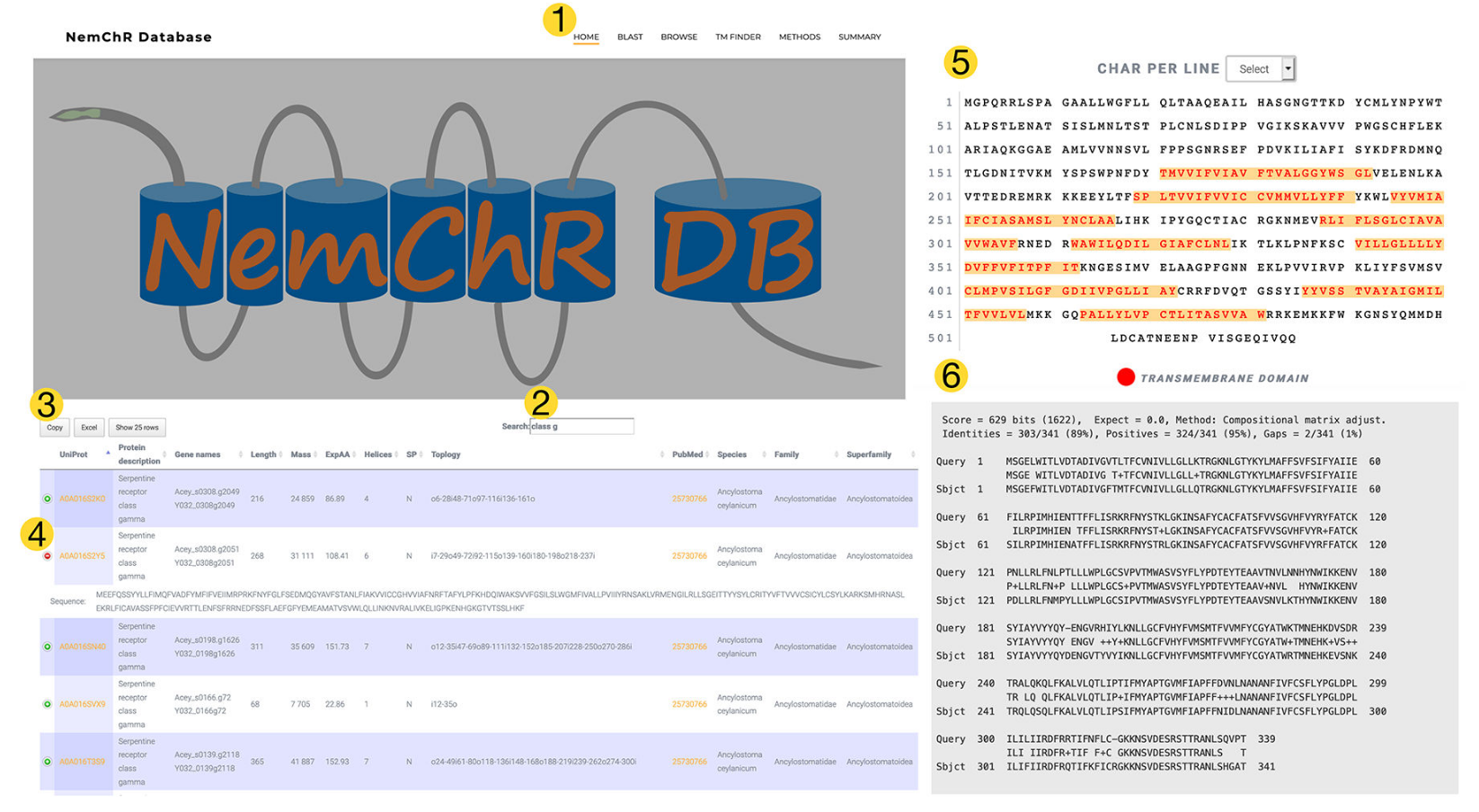
We have shown that these chemoreceptors are upregulated during host seeking stages in hookworm and have mined our chemoreceptor database to identify chemoreceptors that are upregulated in H. bacteriophora.

Additionally, we have identified key signatures of permissive hosts that tune the available host range for parasitic nematodes like hookworm.
Lab Website: www.ohalloranlab.net
Sample Publications
McKean E, Grill E, Choi Y-J, Mitreva, M, O'Halloran DM, Hawdon JM (2024). Altered larval activation response associated with multidrug resistance in the canine hookworm Ancylostoma caninum. Parasitology. 2:1-44. PMID: 38163962
Langeland A, McKean E, O'Halloran DM, Hawdon JM (2023). Immunity mediates host specificity in the human hookworm Ancylostoma ceylanicum. Parasitology. 29:1-24. PMID: 38018393
Langeland A, Grill E, Shetty A, O'Halloran DM, Hawdon JM (2023). Comparative transcriptomics from intestinal cells of permissive and non-permissive hosts during Ancylostoma ceylanicum infection reveals unique signatures of protection and host specificity. Parasitology. 8;1-46. PMID: 36883013
O’Halloran, DM (2021). Database of glutamate gated chloride (GluCl) subunits across 125 nematode species: patterns of gene accretion and sequence diversification. G3 Genes Genomes Genetics. 4;12(2). PMID: 35100348
O’Halloran, DM (2021). CRISPR-PN2: a flexible and genome aware platform for diverse CRISPR experiments in parasitic nematodes. Biotechniques. 71(3):495-498. PMID: 34420406
Langeland, A, Jetter, H and O’Halloran, DM (2021). The diversity of ABC transporter genes across the Phylum Nematoda. Parasitology International. 83, 102357. PMID: 33901678
Motaher, R, Grill, E, McKean, E, Kenney, E, Eleftherianos, I, Hawdon, JM & O’Halloran, DM (2021). Chemogenomic approach to identifying nematode chemoreceptor drug targets in the entomopathogenic nematode Heterorhabditis bacteriophora. Computational Biology and Chemistry. 92(107464). PMID: 33667976
Langeland, A, Hawdon, JM and O’Halloran, DM (2020). NemChR-DB: a database of parasitic nematode chemosensory G-Protein Coupled Receptors. International Journal for Parasitology, S0020-7519(20):30314-3. PMID: 33275943
Bernot, JP, Rudy, G, Erickson, P, Ratnappan, R, Haile, M, Rosa, BA, Mitreva, M, O’Halloran, DM and Hawdon, JM (2020). Transcriptomic analysis of hookworm Ancylostoma ceylanicum life cycle stages reveals changes in GPCR diversity associated with the onset of parasitism. International Journal for Parasitology, 50(8):603-610. PMID: 32592811
Kitchen, S, Ratnappan, R, Han, S, Leasure, C, Grill, E, Iqbal, Z, Granger, O, O’Halloran, DM & Hawdon, JM (2019). Isolation and characterization of a naturally occurring multidrug resistant strain of the canine hookworm Ancylostoma caninum. International Journal for Parasitology, 49(5): 397-406. PMID: 30771359
Sharma, V, Roy, S, Sekler, I & O’Halloran, DM (2017). The NCLX-type Na+/Ca2+ exchanger NCX-9 is required for patterning of neural circuits in Caenorhabditis elegans. Journal of Biological Chemistry, 292(13): 5364–5377. PMID: 28196860
O’Halloran, DM, Altshuler-Keylin, S, Zhang, X, He, C, Morales-Phan, Yu, Y, Kaye, J, Brueggeman, C, Chen, T & L’Etoile, ND (2017). Contribution of the cyclic nucleotide gated channel subunit, CNG-3, to olfactory plasticity in Caenorhabditis elegans. Nature Scientific Reports, 7(1):169. PMID: 28279024
- BSc Maynooth University 2000, PhD Maynooth University 2004
- Postdoc UC Davis 2005-2012
BISC 2220- Developmental Neurobiology
BISC 2320- Neural Circuits and Behavior

